아 그 파일 관련 기능은 다 뺐음. 그리고 이식 하긴 했는데 다른데서 개고생함… ㅋㅋㅋㅋㅋ 그래요 개고생이 끼어야 코딩이지… 한번에 되면 그게 이상한거지…
Cutter
그… 왜 Searcher에서 라디오버튼 선택하면 값 전송하는 건 다들 아실거고… 이걸 Cutter에도 적용해야 한다. 물론 Finder에도 적용할거다. 이게 왜 필요하냐면 입력 수단이 라디오버튼이기 때문. 그리고 라디오버튼에 각각 딸려오는 요소가 있어서, 연결된 라디오버튼이 선택되지 않았을 때 비활성화도 시켜줘야 한다.
이거 근데 Finder 가면서 소스 갈아엎음. ㅇㅇ
function activate() {
const checked_input = document.querySelectorAll('#typing')
const textarea = document.querySelectorAll('textarea')
for (let i = 0; i < checked_input.length; i++) {
if (checked_input[i].checked == true) {
textarea[i].disabled = false;
textarea[i].focus();
} else {
textarea[i].disabled = true;
}
}
}
function upfasta() {
const checked_fasta = document.querySelectorAll('#FASTA')
const upload_fasta = document.querySelectorAll('#input-FASTA')
for (let i = 0;i < checked_fasta.length;i++) {
if (checked_fasta[i].checked == true) {
upload_fasta[i].disabled = false;
}
else {
upload_fasta.disabled = true;
}
}
}
function upgen() {
const checked_gen = document.querySelectorAll('#Genbank')
const upload_gen = document.querySelectorAll('#input-Genbank')
for (let i = 0;i < checked_fasta.length;i++) {
if (checked_gen[i].checked == true) {
upload_gen[i].disabled = false;
}
else {
upload_gen.disabled = true;
}
}
}소스가 왜 바꼈냐면 Finder 이식하면서 직접 입력/FASTA/Genbank가 아니라 커터/파인더로 나눴기 때문이다.
아무튼 Cutter에도 Searcher에 들어가는 라디오버튼이 일부 있다. NEB filter와 cut별 필터가 그거. 그니까 이번에 Ajax를 통해서 보내는 값은
- 입력한 시퀀스
- NEB filter 선택값
- Cut 엔딩 선택값
이렇게 세 개다.
function cutFilter() {
const NEB_filter = document.getElementsByName('option_NEB');
const cut_filter = document.getElementsByName('option_cut');
keyword_val = document.getElementById('keywd').value;
for (let i = 0; i < NEB_filter.length; i++) {
if (NEB_filter[i].checked == true) {
NEB_filter_val = NEB_filter[i].value;
}
}
for (let i = 0; i < cut_filter.length; i++) {
if (cut_filter[i].checked == true) {
cut_filter_val = cut_filter[i].value
}
}
}그리고 필터값과 달리 시퀀스는 이미 직접 입력하겠다고 라디오버튼으로 정한거라 걍 텍스트에리어 값을 보내면 된다.
$.ajax({
type: "POST",
url: "/cutter",
data: {
NEB_give: NEB_filter_val,
cut_give: cut_filter_val,
sequence_give: sequence
},
success: function (response) {
alert(response['msg'])
}
})자 Ajax 드가즈아!!!
app.py
def enz_cutter():
enzyme_table = pd.read_csv('/home/koreanraichu/restriction_merge.csv')
NEB_receive = request.form['NEB_give']
cut_receive = request.form['cut_give']
sequence_receive = request.form['sequence_give']
year = datetime.today().year
month = datetime.today().month
day = datetime.today().day
class RE_treatment:
def RE_wildcard(self, before_seq):
self.before_seq = before_seq
before_seq = before_seq.replace("N", ".")
return before_seq
# Wildcard: 시퀀스 데이터에 N이 있을 경우 Wildcard로 바꾼다.
def RE_or(self, before_seq):
self.before_seq = before_seq
if "B" in before_seq:
before_seq = before_seq.replace("B", "[CGT]")
elif "D" in before_seq:
before_seq = before_seq.replace("D", "[AGT]")
elif "H" in before_seq:
before_seq = before_seq.replace("H", "[ACT]")
elif "K" in before_seq:
before_seq = before_seq.replace("K", "[GT]")
elif "M" in before_seq:
before_seq = before_seq.replace("M", "[AC]")
elif "R" in before_seq:
before_seq = before_seq.replace("R", "[AG]")
elif "S" in before_seq:
before_seq = before_seq.replace("S", "[CG]")
elif "V" in before_seq:
before_seq = before_seq.replace("V", "[ACG]")
elif "W" in before_seq:
before_seq = before_seq.replace("W", "[AT]")
elif "Y" in before_seq:
before_seq = before_seq.replace("Y", "[CT]")
return before_seq
if cut_receive == 'sticky':
enzyme_table = enzyme_table[enzyme_table['cut_feature'] == 'Sticky']
enzyme_table.reset_index(inplace=True)
elif cut_receive == 'blunt':
enzyme_table = enzyme_table[enzyme_table['cut_feature'] == 'Blunt']
enzyme_table.reset_index(inplace=True)
elif cut_receive == 'nicked':
enzyme_table = enzyme_table[enzyme_table['cut_feature'] == 'Nicked']
enzyme_table.reset_index(inplace=True)
else:
cut_receive = "All feature"
pass
if NEB_receive == "NEB":
enzyme_table = enzyme_table[enzyme_table['NEB_sell'] == 'Yes']
enzyme_table.reset_index(inplace=True)
else:
NEB_receive = "All selling enzyme"
pass
sequence_name = "Typed sequence"
sequence_description = "Typed sequence"
# 여기가 위치 관련 함수입니다.
def convert(a):
RE = RE_treatment()
if "N" in res_find:
res_find_after = RE.RE_wildcard(res_find)
elif "B" in res_find or "D" in res_find or "H" in res_find or "K" in res_find or "M" in res_find or "R" in res_find or "S" in res_find or "V" in res_find or "W" in res_find or "Y" in res_find:
res_find_after = RE.RE_or(res_find)
return res_find_after
count = 0
count_nocut = 0
once_cut_list = []
two_cut_list = []
multi_cut_list = []
no_cut_list = []
result_msg_list = []
# 변수와 리스트(크게 건들 일 없음)
head_msg = "=====Sequence information=====\nSequence name: {0} | Sequence length: {1}bp \nSequence description: {" \
"2}\n=====Running information======\nFilter selected: {3} | {4} \nRestriction enzyme which cuts this " \
"\n =====Result=====\n".format(sequence_name, len(sequence_receive), sequence_description, cut_receive,
NEB_receive)
for i in range(len(enzyme_table)):
enzyme = enzyme_table['Enzyme'][i]
feature = enzyme_table['cut_feature'][i]
res_find = enzyme_table['sequence'][i]
res_find = str(res_find)
res_find_before = str(res_find)
while True:
if "N" in res_find:
res_find = str(convert(res_find))
elif "B" in res_find or "D" in res_find or "H" in res_find or "K" in res_find or "M" in res_find or "R" in res_find or "S" in res_find:
res_find = str(convert(res_find))
else:
break
# 정규식 처리
Findall = re.findall(res_find, sequence_receive)
if Findall:
res_loc_list = []
count += 1
site_count = len(Findall)
locs = re.finditer(res_find, sequence_receive)
for i in locs:
loc = i.start()
res_loc_list.append(str(loc + 1))
if site_count == 1:
once_cut_list.append(enzyme)
elif site_count == 2:
two_cut_list.append(enzyme)
else:
multi_cut_list.append(enzyme)
res_loc_list = ', '.join(res_loc_list)
result_msg = ("Enzyme: {0} | Sequence: {1} | Cut feature: {2} | {3} times cut \nWhere(bp): {4} \n".format(
enzyme,
res_find_before,
feature,
site_count,
res_loc_list))
result_msg_list.append(result_msg)
else:
count += 0
count_nocut += 1
no_cut_list.append(enzyme)
once_cut_list = ', '.join(once_cut_list)
two_cut_list = ', '.join(two_cut_list)
multi_cut_list = ', '.join(multi_cut_list)
no_cut_list = ', '.join(no_cut_list)
# 출력부
end_msg = ("Total: {0} enzymes cut input sequence, {1} enzymes never cut this sequence. \nEnzymes no cut this "
"sequence: {2} \nEnzymes cut this sequence once: {3} \nEnzymes cut this sequence twice: {4} "
"\nEnzymes cut this sequence multiple: {5} \n".format(count,
count_nocut,
no_cut_list, once_cut_list,
two_cut_list, multi_cut_list))
return jsonify({'head_give': head_msg,'result_give':result_msg_list,'end_give':end_msg})script.js
function cutFilter() {
const checked_input = document.querySelectorAll('#typing')
const textarea = document.querySelectorAll('textarea')
const checked_fasta = document.querySelectorAll('#FASTA')
const checked_gen = document.querySelectorAll('#Genbank')
const NEB_filter = document.getElementsByName('option_NEB');
const cut_filter = document.getElementsByName('option_cut');
let sequence = '';
if (checked_input[0].checked == true) {
sequence = textarea[0].value;
}
else {
sequence = 'not supported'
}
for (let i = 0; i < NEB_filter.length; i++) {
if (NEB_filter[i].checked == true) {
NEB_filter_val = NEB_filter[i].value;
}
}
for (let i = 0; i < cut_filter.length; i++) {
if (cut_filter[i].checked == true) {
cut_filter_val = cut_filter[i].value
}
}
$.ajax({
type: "POST",
url: "/cutter",
data: {
NEB_give: NEB_filter_val,
cut_give: cut_filter_val,
sequence_give: sequence
},
success: function (response) {
let head_msg = response['head_give']
let result_msg = response['result_give']
let end_message = response['end_give']
let result = document.getElementById('cut_result');
result.innerHTML = head_msg;
for (let i = 0; i < result_msg.length; i++) {
result.innerHTML = result.innerHTML.concat("\n", result_msg[i])
}
result.innerHTML = result.innerHTML.concat("\n", end_message)
}
})
}참고로 얘는 로직 이식할 때 개고생했다. 값은 생각보다 수월하게 가져왔는데… Flask에 갖다놨더니 애가 전역변수를 인식을 못해서 함수로 뺐던거 다시 코드에 넣었다.
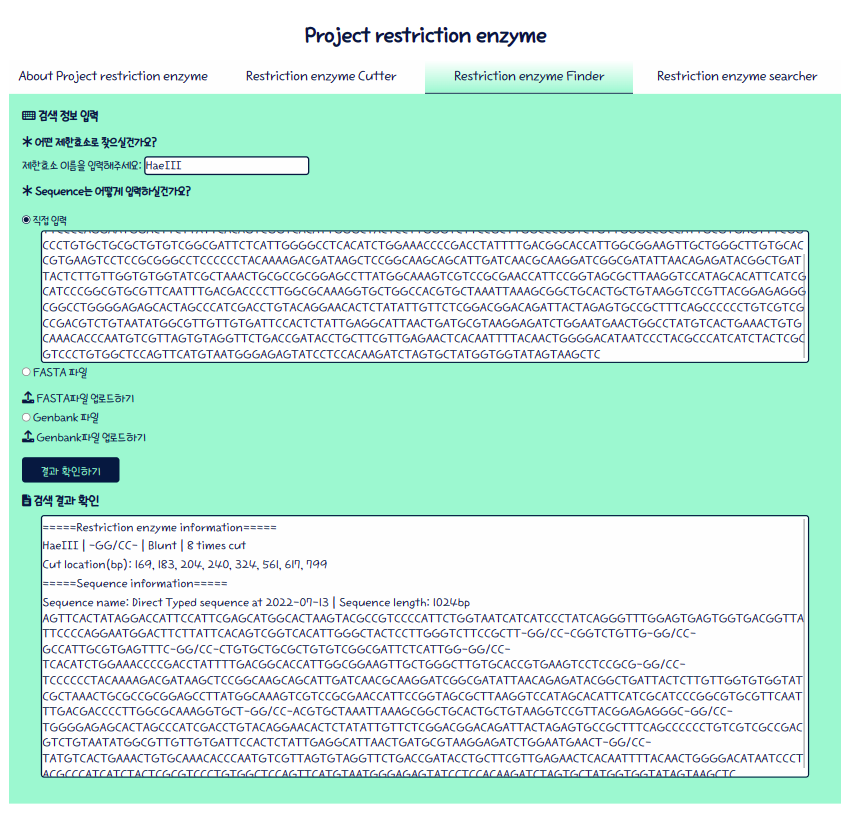
Finder
얘는 로직 이식은 생각보다 수월한데 반대로 JS쪽에서 개고생했다. 뭐가 불만인지 라디오버튼을 선택해도 세상에 활성이 안되잖어… 그래서 커터/파인더별로 묶었다. 용도별로 묶었더니 checked_input[i]가 체크 된 건 인식하는데 i번째 오브젝트 disabled가 안 풀렸거든…
const checked_input = document.querySelectorAll('#typing');
const input_enz = document.getElementsByName('enzyme');
const textarea = document.querySelectorAll('textarea');
const checked_fasta = document.querySelectorAll('#FASTA');
const checked_gen = document.querySelectorAll('#Genbank');
const NEB_filter = document.getElementsByName('option_NEB');
const cut_filter = document.getElementsByName('option_cut');
let sequence = '';
if (checked_input[1].checked == true) {
sequence = textarea[1].value;
}
for (let i = 0; i < NEB_filter.length; i++) {
if (NEB_filter[i].checked == true) {
NEB_filter_val = NEB_filter[i].value;
}
}
for (let i = 0; i < cut_filter.length; i++) {
if (cut_filter[i].checked == true) {
cut_filter_val = cut_filter[i].value
}
}그것만 빼면 괜찮아서 입력 갈아엎고 스무th하게… 가긴 무슨… Finder는 이 효소가 이 시퀀스를 자르는가를 보는거라 저 필터들이 아예 없다. 그래서 저기 라디오버튼 걸려있는 필터들을 죄다 빼야 한다. 그러니까, 저기 있는 for문 두 개 얘기한거다.
function findFilter() {
const input_enz = document.getElementById('enzyme').value;
const textarea = document.querySelectorAll('textarea');
const checked_fasta = document.querySelectorAll('#FASTA');
const checked_gen = document.querySelectorAll('#Genbank');
let sequence = textarea[2].value
console.log(input_enz,sequence)
$.ajax({
type: "POST",
url: "/finder",
data: {
sequence_give: sequence,
enzyme_give: input_enz
},
success: function (response) {
console.log(response)
}
})
}그래서 입력 박스 두 개의 값만 전달하면 된다. 참 쉽죠?
참고로 저게 다라 이식 자체는 싱겁게 끝났다. 전역변수 없는건 커터덕에 알아서 바로 수정했고… 근데 format땜시 {}준걸 왜 니가 나눠놓고 인식을 못하냐 파이참…
app.py
@app.route('/finder', methods=['POST'])
def enz_finder():
enzyme_table = pd.read_csv('/home/koreanraichu/restriction_merge.csv')
enzyme_receive = request.form['enzyme_give']
sequence_receive = request.form['sequence_give']
year = str(datetime.today().year)
month = str(datetime.today().month).zfill(2)
day = str(datetime.today().day)
sequence_name = "Direct Typed sequence at {0}-{1}-{2}".format(year, month, day)
sequence_description = "Direct Typed sequence at {0}-{1}-{2}".format(year, month, day)
class RE_treatment:
def RE_wildcard(self, before_seq):
self.before_seq = before_seq
before_seq = before_seq.replace("N", ".")
return before_seq
# Wildcard: 시퀀스 데이터에 N이 있을 경우 Wildcard로 바꾼다.
def RE_or(self, before_seq):
self.before_seq = before_seq
if "B" in before_seq:
before_seq = before_seq.replace("B", "[CGT]")
elif "D" in before_seq:
before_seq = before_seq.replace("D", "[AGT]")
elif "H" in before_seq:
before_seq = before_seq.replace("H", "[ACT]")
elif "K" in before_seq:
before_seq = before_seq.replace("K", "[GT]")
elif "M" in before_seq:
before_seq = before_seq.replace("M", "[AC]")
elif "R" in before_seq:
before_seq = before_seq.replace("R", "[AG]")
elif "S" in before_seq:
before_seq = before_seq.replace("S", "[CG]")
elif "V" in before_seq:
before_seq = before_seq.replace("V", "[ACG]")
elif "W" in before_seq:
before_seq = before_seq.replace("W", "[AT]")
elif "Y" in before_seq:
before_seq = before_seq.replace("Y", "[CT]")
return before_seq
# 여기가 위치 관련 함수입니다.
def convert(a):
RE = RE_treatment()
if "N" in res_find:
res_find_after = RE.RE_wildcard(res_find)
elif "B" in res_find or "D" in res_find or "H" in res_find or "K" in res_find or "M" in res_find or "R" in res_find or "S" in res_find or "V" in res_find or "W" in res_find or "Y" in res_find:
res_find_after = RE.RE_or(res_find)
return res_find_after
res_find = enzyme_table.sequence[(enzyme_table['Enzyme'] == enzyme_receive)]
res_find = res_find.to_string(index=False)
res_find = res_find.upper()
res_find = str(res_find)
while True:
if "N" in res_find:
res_find = str(convert(res_find))
elif "B" in res_find or "D" in res_find or "H" in res_find or "K" in res_find or "M" in res_find or "R" in res_find or "S" in res_find:
res_find = str(convert(res_find))
else:
break
# 정규식 처리
res_site = enzyme_table.restriction_site[(enzyme_table['Enzyme'] == enzyme_receive)]
res_site = res_site.to_string(index=False)
res_site = res_site.upper()
res_site = str(res_site)
# 자르는 시퀀스 처리
cut_feature = enzyme_table.cut_feature[(enzyme_table['Enzyme'] == enzyme_receive)]
cut_feature = cut_feature.to_string(index=False)
cut_feature = str(cut_feature)
# blunt or sticky(나중에 저장 기능 추가할 때 넣을 예정입니다)
Findall = re.findall(res_find, sequence_receive)
if Findall:
site_count = 0
res_loc = 0
res_loc_list = []
locs = re.finditer(res_find,sequence_receive)
for i in locs:
loc = i.start()
res_loc_list.append(str(loc + 1))
cut_count = len(Findall)
if len(set(Findall)) > 1:
for i in Findall:
if i in sequence:
sequence = sequence.replace(i, "-" + i + "-")
else:
sequence = sequence_receive.replace(res_find, res_site)
res_loc_list = ', '.join(res_loc_list)
head_msg = "=====Restriction enzyme information=====\n{0} | {1} | {2} | {3} times cut \nCut location(bp): {4} \n=====Sequence information=====\nSequence name: {5} | Sequence length: {6}bp \n{7}".format(
enzyme_receive,
res_site,
cut_feature,
cut_count,
res_loc_list, sequence_name,
len(sequence),
sequence)
elif not Findall:
head_msg = "=====Restriction enzyme information=====\n{0} | {1} | {2} | 0 times cut \nThis restricion enzyme " \
"no cut this sequence. \n=====Sequence information=====\nSequence name: {3} | Sequence length: {4}bp \n{5}".format(enzyme_receive, res_site,
cut_feature, sequence_name,
len(sequence_receive),
sequence_receive)
else:
head_msg = "{0} \nThis restriction enzyme not entried in database. ".format(enzyme_receive)
# DB에 효소가 없을 때
return jsonify({'head_give': head_msg})script.js
function findFilter() {
const input_enz = document.getElementById('enzyme').value;
const textarea = document.querySelectorAll('textarea');
const checked_fasta = document.querySelectorAll('#FASTA');
const checked_gen = document.querySelectorAll('#Genbank');
let sequence = textarea[2].value
$.ajax({
type: "POST",
url: "/finder",
data: {
sequence_give: sequence,
enzyme_give: input_enz
},
success: function (response) {
console.log(response)
let msg = response['head_give']
let result = document.getElementById('find_result');
result.innerHTML = msg;
}
})
}주는게 적다=받는게 적다=profit?
완성본



Reply